Prepared by Tonya Amen, Ph.D. on behalf of the Texas Angus Association for the September 2017 Newsletter.
I was asked to write this month’s article about “Single-Step”, but I suspect the real question is what happened with AAA EPDs on July 7? It may be “Single-Step” that harnessed the most press and attracted most of the attention but there were several other major changes made to the evaluation at the same time. In the following paragraphs, I’ll give a brief description of single step and the advantages of moving to the new method and then describe the other changes that were made to the evaluation and their apparent impact.
What is Single-Step?
A new, more streamlined method for incorporating genomic results into national cattle evaluations has been widely discussed not only in Angus publications, but also in the broader industry at numerous events and conferences for the last couple of years. You’ve likely heard it referred to either as “Single-Step” or as “BOLT” (BOLT is the name of a software program that some breeds will use to launch their single-step evaluation later this year; AAA uses University of Georgia software).
Prior to July 7, AAA GE-EPDs were generated using a multi-step approach, which necessitated semi-regular recalibrations (Figure 1). To begin the calibration process, all available pedigree and performance information was used in a “classic” genetic evaluation (A), which resulted in what were essentially “old-school” EPDS without genomics (B). These “classic” EBVs were then used along with genomic results (SNP) to estimate the effects of those SNPS on the various EPD traits (C). These SNP effects were then added together to arrive at a molecular breeding value (MBV). The last step in each recalibration was calculate new correlations and heritabilities (REML in D, below). Finally, the pedigree, performance data and MBVs were run together each week to produce GE-EPDs (E). The results from this method were state-of the art at the time, and provided an improvement in the accuracy of EPDs which were previously estimated with only pedigree and performance information. But, the research required for each calibration was lengthy and costly, plus it typically left some uncertainty with breeders awaiting the timing of its release.
With the migration to the Single-Step method (Figure 2), a more streamlined process was established. Each week, pedigrees, performance information and genomic results will all be used in a single step to generate GE-EPDs. Many articles have been written about the benefits of this computational advancement (see Hermel and AGI publications in the references, to name just a couple), so I’ll summarize the benefits just briefly
- Eliminates the need for recalibration. This saves time, money and, equally important, eliminates the marketing and communication disruption caused by the sometimes large changes resulting from each recalibration.
- Makes room in the model for other traits. This is a tough one to grasp unless you’re accustomed to working matrix algebra and solving equations using large matrices. For each trait that is added to a model (genomic results were handled like another correlated trait in the multi-step method) many more equations had to be solved to generate results necessary for EPDs. Computing power and time restraints associated with a weekly evaluation were a very real concerns.
- Explains the relationship between animals better. In our early, basic genetics or biology classes, most of us were taught that, on average, full-sibs share 50% of their genetic material and that, on average, grandparents and grand-progeny should share about 25%. Of course, the key here is “on average”. These averages were the standard by pedigree relationships were described prior to single-step. In theory, full-sibs could share 0-100% of their genetic material (in practice, this doesn’t happen due to linkage and some other things beyond the scope of this article). However, using single step can refine the relationship between animals beyond the standard ½, ¼, etc. See the Decker (2015) article in the references for more in-depth discussion of this.
- More descriptive accuracies. Using the multi-step approach, a genomic result was a genomic result accuracy-wise. Using single-step, those breeders who have invested in genomics and performance data collection should recognize higher accuracy values for their animals that those who haven’t.
What Else Changed?
- Due to the popularity of the $Beef index, anything that is done to the underlying carcass or growth EPDs is going to get a lot of attention and I suspect most of you noticed the changes that were made to the carcass model. Migration to single-step freed up some space in the model to add weaning weight and fat thickness and their associated correlations. This should help with some selection bias that may have occurred due to animals with weaning records being culled before yearling scan-weights were taken and more fully represent potential differences in the growth curve of animals.
- Additionally, birth weight and weaning weight are now reciprocally used in each other’s EPD computation. This will allow Birth Weights to be used in EPDs calculation before the submission of weaning weights and will also help improve the accuracy of weaning weight EPD by allowing BW to be used as a correlated trait.
- Any time substantive changes are made to NCE models, the heritabilities of the traits and correlations between them must updated. Changing heritability does not impact the rank of animals, but it does impact the number of progeny records that are required to achieve a given level of accuracy. Adding correlated traits and/or changing the magnitude of genetic correlations between traits could have an impact on EPDs.
- As is customary on a biannual basis in the AAA evaluation, the economic assumptions were updated.
- Associated tables had to be updated. Breed averages shifted and the percentile rank table changed (somewhat significantly for some traits). You’ll need to “recalibrate” your internal scale for what is top 5% vs top 50% and be sure to help your customers with the new scale.
Summary
Though I’ve heard a lot of cussing and discussing genomics, the July 7 release represented a very major overhaul of several of the highest profile EPDs. In addition to changing the way that genomic results are accounted for in the evaluation, traits were added to several models, new correlations and heritabilities were released, the economic assumptions behind the $-Value Indices were updated AND the number of genomic results entering the evaluation had more than tripled since the previous recalibration. That’s a lot to try to absorb and understand (not to mention explain to your customers).
The good news is that the evaluations should be less dramatic going forward, with small changes resulting from performance data and weekly accumulation of genomic results. There are winners and losers each time a change this substantial is made, so I won’t try to minimize the significance for those who may be disappointed in the changes. However, recognize that there were a lot of things at play in the changes that occurred, and teasing out what caused the changes in specific animals is a difficult (if not impossible) mission. Keep in mind what you’ve come to know about EPDs:
- Performance data is more important than ever, submit as much as you can from proper-formed contemporary groups
- Genomic results are a useful for adding accuracy to young animals and those with lower accuracies – they are not a replacement for performance data.
- EPDs are the best estimate of an animal’s genetic value as a parent for the traits that are measured – use them.
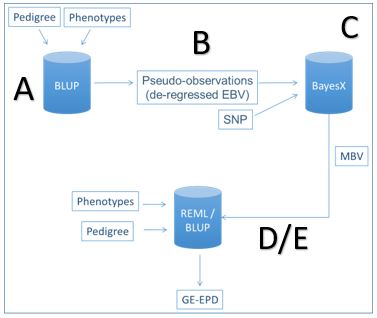 Figure 1 – Multi-Step Method for GE-EPD Generation. From Lourenco, 2017.
Figure 1 – Multi-Step Method for GE-EPD Generation. From Lourenco, 2017.
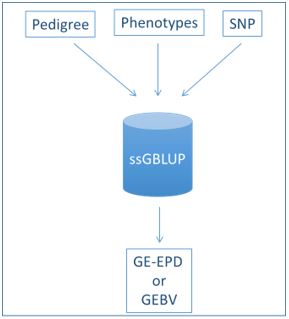 Figure 2 – Single-Step Method for GE-EPD Generation. From Lourenco 2017.
Figure 2 – Single-Step Method for GE-EPD Generation. From Lourenco 2017.
Decker, J.E. (2015). The Random Shuffle of Genes: Putting the E in EPD. Available: http://articles.extension.org/pages/72650/the-random-shuffle-of-genes:-putting-the-e-in-epd
AGI (2017). Single-Step Genetic Evaluation – Common Questions. Available: https://www.angus.org/AGI/Single-stepGeneticEvaluation%C2%ADCommonQuestions.pdf
Hermel, S.R. (2017). Angus to Convert to Single-Step July 7. Proceedings Beef Improvement Federation Conference, Athens, GA. Available: http://www.bifconference.com/bif2017/summaries/GGP-StephenMiller.htm
Lourenco, D. et. al (2017). The Promise of Genomics for Beef Improvement. Proceedings Beef Improvement Federation Conference, Athens, GA.http://www.bifconference.com/bif2017/documents/GS1-Lourenco-slides.pdf


